A randomized controlled trial of heterologous ChAdOx1 nCoV-19 and recombinant subunit vaccine MVC-COV1901 against COVID-19

Study design
This was an investigator-initiated, single-blinded, 1:1 randomized vaccine clinical study, designed to assess the reactogenicity and immunogenicity of heterologous prime-boost immunization with ChAdOx1 (AZD1222) followed by the subunit vaccine MVC-COV1901, compared with the homologous immunization with two doses of ChAdOx1 (Fig. 1). Participants were healthy adults without severe disorders at the age of 20–70 years who have had their first dose of the ChAdOx1 vaccine. There was no TTS or other serious adverse events following the first ChAdOx1 vaccination in all participants. For female participants, they must be either of non-childbearing potential (i.e., surgically sterilized or one year post-menopausal) or, if of childbearing potential, be abstinent or agree to use medically effective contraception on enrollment continuously until 90 days after boost immunization of study intervention. A negative pregnancy test was required before enrollment.
The primary objective of this trial was to determine if the immune response (neutralizing antibody titer at day 28 after the booster dose) of heterologous group was non-inferior to that observed in the homologous group (Supplementary Note 1). The non-inferiority study design was based on the immuno-bridging standards in granting Emergency Use Authorization for COVID-19 vaccine (including MVC-COV1901) by Taiwan FDA (https://www.fda.gov.tw)20. The immuno-bridging success criteria was the lower limit of the 2-sided 95% confidence interval (CI) for geometric mean titer (GMT) ratio >0.6720.
The study was conducted in a single institute in Chang Gung Memorial Hospital, Linko branch, in Taiwan. After receiving the treatment, the participants remained in the study for 168 days following booster vaccination.
Written informed consent was obtained from all participants, and the trial is being done in accordance with the principles of the Declaration of Helsinki and Good Clinical Practice. This study was approved by the Taiwan Food and Drug Administration and the ethics committee at Chang Gung Medical Foundation (Taiwan). The study was registered in ClinicalTrials.gov with ID NCT05054621 and the protocol in detail is available in Supplementary information.
Outcomes
For the study primary objective, humoral immunogenicity including serologic neutralizing antibody titers against SARS-CoV-2 and serological quantification of binding antibody to SARS-CoV-2 antigen was assessed during the duration of the study at baseline and after booster vaccination at day 10 ± 3, day 28 ± 3, day 56 ± 3 and day 168 ± 7. The SARS-CoV-2 antigen specific B cell and T cell frequencies were assessed in day 0, day 10 ± 3 and day 28 ± 3 after booster vaccination. Safety was assessed during the duration of the study. The solicited adverse events (AEs) occurring locally or systemically were assessed for 7 days following each vaccination from day 0 through day 7. Unsolicited AEs were recorded for 28 days after the boost dose. Serious AEs (SAEs) were recorded from signing of the informed consent form through day 168. Adverse events of special interest (AESIs) were recorded from the booster vaccination through day 168.
Randomization and blinding
All eligible participants were 1:1 randomly assigned to receive a single dose of either the same vaccine as their prime dose ChAdOx1 (homologous group) or the Medigen COVID-19 vaccine MVC-COV1901 (heterologous group). Stratified randomization was used based on the intervals between prime and boost vaccination. Participants were stratified according to the prime-boost intervals of 4–6 weeks and 8–10 weeks, respectively, with equal-sized strata. Randomization was applied to each stratum and the random number list was generated by an independent study statistician using SAS software.
The treatment phase was conducted in a single-blinded fashion such that the participants were masked to the vaccine received but not to the prime-boost interval. Clinical staffs who involved in the vaccine delivery were aware of which vaccine the participant received, but the participant remained blinded by preparing the vaccine out of sight and applying a masking tape over the vaccine syringe. Laboratory staffs were also blinded to the vaccine the participant received, which may minimize the evaluation bias from the knowledge about the treatment assignment of the participant.
Surrogate neutralizing titers by ELISA method
All serum samples were analyzed by the SARS-CoV-2 antibody ELISA kit according to the manufacturers’ instructions (MeDiPro, Taiwan)21,22. MeDiPro was a Taiwan FDA-approved kit for quantifying Spike S1-and receptor-binding domain (RBD)-binding antibodies which were surrogates of live virus neutralization titers with high correlation21,22. With the cutoff of <34.47 IU/mL defining negative result, the sensitivity and specificity of the test was 92.2% (95% Cl, 84.0%–96.4%) and 93% (95% Cl, 81.4%–97.6%), respectively.
Live virus neutralization assay
The neutralization assay following the standard protocol of a plaque reduction neutralization test was performed on the serum samples collected at day 28 after booster dose of vaccination. Vero cells were regularly maintained in minimal essential medium (MEM) supplemented with 10% (v/v) fetal bovine serum. Wild type virus (Wuhan strain) and Delta variant of SARS-CoV-2 were propagated in Vero cells in MEM. Serum samples were inactivated at 56 °C for 30 min before use. Serum were two-fold diluted serially and were mixed with equal volumes of SARS-CoV-2 suspension containing 100 folds of the median tissue culture infectious dose. The mixture was incubated for 2 h at 37 °C, and then an equal volume of suspended Vero E6 cells (approximately 30,000 cells/well) was added to each well. Following incubation for 1 week at 37 °C, cells were fixed with 5% glutaraldehyde and stained with 0.1% crystal violet. Serum neutralization titers were calculated and expressed as the reciprocals of the highest serum dilution that inhibits cytopathic effects.
PBMC preparation
PBMCs were separated from heparinized blood by density gradient centrifugation using lymphoprep (Stemcell Technologies, Canada) for 20 min, 800 g at 20 °C. The PBMC band was collected, transferred to a sterile conical tube pre-filled with RPMI medium and centrifuged for 10 min, 720 g. The supernatant was discarded, the cell pellet was resuspended in RPMI medium containing 10% fetal bovine serum and centrifuged for 10 min, 400 g. Following this centrifugation, the supernatant was discarded, and the cell pellet was resuspended in RPMI medium containing 10% fetal bovine serum. An aliquot of cell suspension was used for counting and viability assessment.
Ex vivo enzyme-linked immunospot for detection of antibody-secreting B cell response
Sterile, clear 96-well filter plate with 0.45 μm pore size hydrophobic PVDF membrane (MAIPS4510, Millipore, United States) were coated with 100 μL of 15 μg/mL Wuhan-Hu-1 strain SARS-CoV-2 spike trimer diluted in carbonate buffer or carbonate buffer only as negative control or polyvalent anti-human Ig’s (Thermo Fisher Scientific, United States) as positive control and incubated overnight at 4 °C. Plates were washed with PBS three times and blocked using RPMI medium containing 10% fetal bovine serum for 1 h at 37 °C. Blocking medium was removed and plates were washed with PBS. 100 μL of PBMC suspension was added at a density of 2 × 105 cells per well for antigen-specific response and 2 × 104 cells per well for total IgG, IgM and IgA response (positive control) and incubated for 18 h at 37 °C. Cell suspension was removed and plates were washed with PBS. 100 μL of 1:5000 diluted alkaline phosphatase conjugated anti-human IgG, IgM or IgA (Calbiochem, United States) was added and incubated for 2 h at room temperature. After washing with PBS, 50 μL of BCIP/NBT-plus substrate (Mabtech, United States) was added and left for 2 to 5 minutes at room temperature. After distinct spots developed, the reaction was stopped using distilled water. Plates were air-dried and spots were measured and counted with automatic ELISpot reader.
Ex vivo interferon-γ enzyme-linked immunospot for detection of cellular response
Interferon-γ ELISpot assay was performed using the human IFN-γ ELISpot basic kit (ALP) (3420-2 A, Mabtech, United States) according to the manufacturer’s protocol with some modifications. Sterile, clear 96-well filter plate with 0.45 μm pore size hydrophobic PVDF membrane (MAIPS4510, Millipore, United States) was coated by 100 μL of 15 μg/mL anti-human IFN-γ monoclonal antibody (1-D1K) (Mabtech, United States) in sterile PBS overnight at 4 °C. Plates were then washed with PBS five times and blocked with 250 μL per well of RPMI medium containing 10% fetal bovine serum for 1 h at 37 °C. After removing the medium, 100 μL of PBMC suspension was added to each well, and SARS-CoV-2 spike S1 subunit or S2 subunit peptide pool (PP003, Sino Biological, China) at a final concentration of 2 μg/ml was added. Control wells of cells incubated with 0.4% DMSO (negative control), or with 10 μg/mL phytohemagglutinin (PHA) (positive control) were also included. After incubation for 18 h at 37 °C, plates were washed with PBS six times and incubated with 100 μL of 1 μg/mL biotinylated anti-human IFN-γ antibody (7-B6-1) (Mabtech, United States) in sterile PBS with 5% BSA for 2 h at room temperature. Plates were then washed with PBS six times and incubated with 50 μL of 1:1000 dilution of Streptavidin-Alkaline phosphatase in sterile PBS for 1 h at room temperature. Plates were then washed with PBS three times and developed with 100 μL of substrate solution (Mabtech, United States) until distinct spots emerged. The color development was terminated by washing plates with distilled water. Plates were air-dried and spots were measured and counted with an automatic ELISpot reader.
Sample size
The primary objective of this trial was to determine if the immune response of heterologous group was non-inferior to that observed in homologous group, and the primary endpoints was neutralizing antibody titer at day 28 after booster vaccination. By assuming the non-inferiority margin was 0.67-fold-difference or −0.401 absolute difference of log GMT between heterologous group and homologous group with the standard deviation 0.66, and the true difference of log GMT was 0, the study needed to recruit 44 evaluable participants per group (total 88 participants) to achieve 80% of power at one-sided 2.5% significance level. According to the missing rate 10% and the stratification in 1:1 for prime-boost 4–6 and 8–10 weeks, 50 participants for each stratum and equally random assigned to each group (25 for heterologous and 25 for homologous group) within strata (total 100 participants) is needed. The mean difference of log GMT was presented with the two-sided 95% CI.
Statistical analysis
Descriptive statistics on continuous measurements included means, medians, standard deviations, and ranges, while categorical data was summarized using frequency counts and percentages. For the immunogenicity endpoints including SARS-CoV-2 neutralizing antibody levels and cell-mediated immune responses, the point estimates were reported with 95% confidence intervals. For the secondary endpoints for comparisons of continuous scale between groups, independent t-test was used. For the comparison of nAb titer changes from baseline between groups of short and long prime-boost intervals, a value of 1 was used to substitute a change value with zero or a value less than zero, so that the information contained in these data was not lost in calculation of geometric mean.
For safety analysis, the number (%) of subjects with AEs was reported. Frequency counts and percentages were also be presented of subjects with serious AEs, AEs leading to withdrawal, AEs by severity and AEs by relationship to study treatment. All other safety measures were analyzed for the safety population.
Evidence of significant interaction was assessed at the 5% level. All analyses were performed using the Statistical Analysis System (SAS) statistical software package, version 9.4. (SAS Institute Inc, Cary, NC) and Graphpad Prism (Version 9.1.1, GraphPad Software, US).
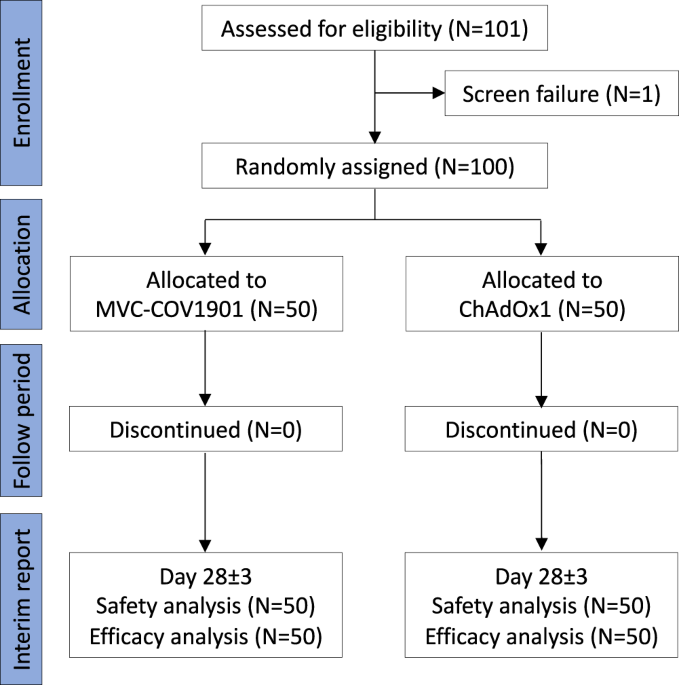
Interim report and enrolment status
The study duration for each participant would be nearly or more than 6 months following the enrolment (visit day −70∼−1, 0, 7, 10 ± 3, 28 ± 3, 56 ± 3 and 168 ± 7, Supplementary Note 1). This analysis was prospectively specified in the protocol (Supplementary Note 1) and reported based on the data collected from all enrolled participants until day 28. As of 01 February 2022, a total of 101 subjects were enrolled in the study. One subject failed screening because he was unable to visit the study site in the scheduled time points. The remaining 100 were followed up, and 0 completed the study when this analysis was reported. The earliest vaccination campaign start date was 15 September 2021.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.


